Introduction
A common challenge in discovery and process chemistry is to find the optimal reaction conditions, including the choice of catalyst and ligand, through the fewest number of experiments. This problem is particularly acute when the reaction space is large and there are limits on the number of experiments that can be performed due to cost or time constraints.
Traditional optimization approaches can be time consuming and costly if knowledge of the system and the initial experiments are far away from the global optimum. Machine learning in the form of Bayesian optimization (BO) proves particularly suited to the challenge of chemical reaction optimization because it works with small datasets and can explore very large reaction spaces.
Several open-source python packages for Bayesian Optimization in chemistry applications have been reported,1–7 however, many are only accessible to scientists with coding expertise.‡ Suppose I was a laboratory scientist and wanted to use my experimental data stored in an electronic lab notebook (ELN) for Bayesian Optimization. How could I go about doing this?
A solution is to use BIOVIA Pipeline Pilot8 to extract and process data from the BIOVIA Notebook9 ELN, run the Bayesian Optimization code, and then update ELN entries with the next round of suggested experiments. By leveraging the capabilities of Pipeline Pilot, we can construct a protocol that utilizes existing components to perform much of the data extraction and manipulation.
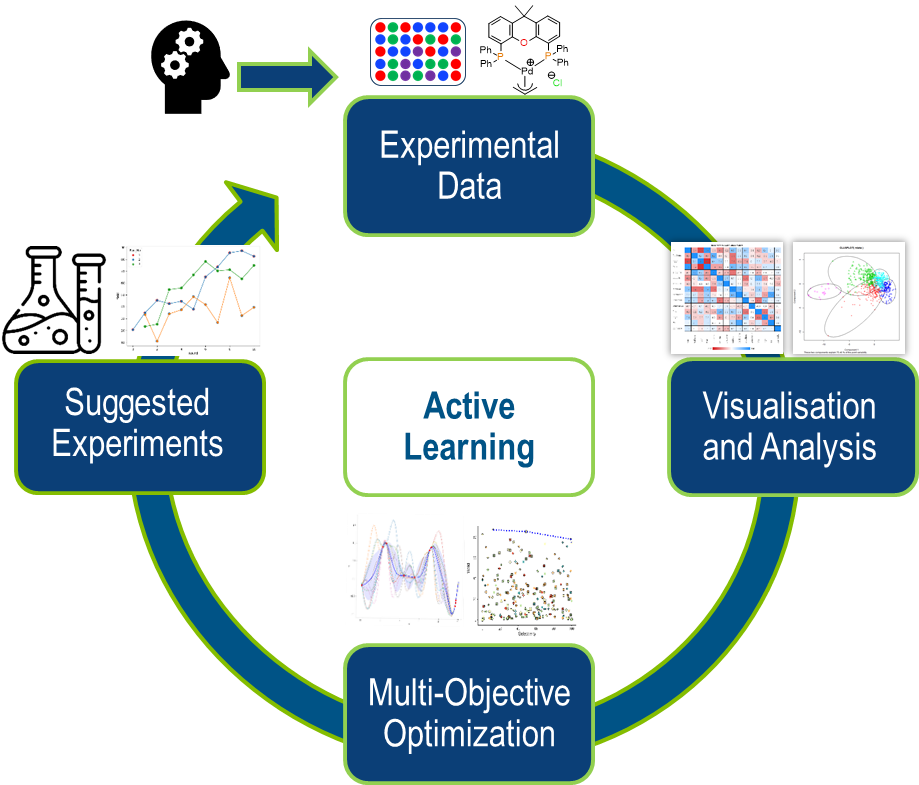
Figure 1: A Bayesian Optimization Workflow
Initial Bayesian Optimization
For this example, we are going to use the EDBO+ Python package as reported by Doyle and co-workers4 and a dataset obtained from a recent publication by Syngenta that explores Bayesian optimization for Ullmann type C-N couplings aiming to maximize reaction yield.10 The Python package is accessed through a Jupyter Notebook component in Pipeline Pilot.
The experimental data for an initial set of eight experiments has been entered into an experiment in the Notebook ELN and one-hot encoding is used for categorical features as implemented in the EDBO+ package.
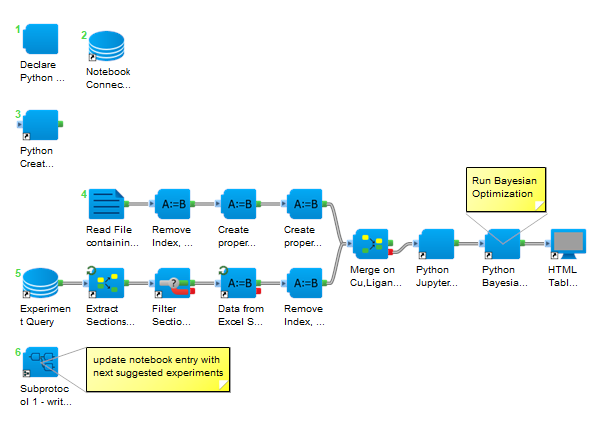
Figure 2: Pipeline Pilot Protocol to run Bayesian Optimization with experiments in BIOVIA Notebook ELN.
The protocol consists of a number of steps:
- Generating an experimental scope based on every combination of the reaction components. In this example that is 138,240 experiments.
- Extracting and cleaning experimental data from the corresponding Notebook experiment before merging with the experimental scope.
- Running Bayesian optimization to suggest the next set of experiments to optimize reaction yield.
- Updating the Notebook entry with the next experiments to run.
The Bayesian Optimization loop can be repeated many times by entering new experimental data into the ELN and re-running the protocol until reactions that deliver the highest yield have been found.
Increased complexity with DFT features
Additional layers of complexity can be built into the Bayesian optimization protocol in Pipeline Pilot by encoding some of the categorical features with DFT derived molecular descriptors. Using SMILES strings as input, we can use Pipeline Pilot to generate 3D coordinates, perform a conformer search and run DFT calculations with existing components in BIOVIA Pipeline Pilot Solvation Chemistry Collection11 or BIOVIA Pipeline Pilot Materials Studio Collection.12 No coding expertise is required to build and run these protocols.
The DFT features can then be used in an updated Bayesian Optimization protocol (Figure 3). Additionally, the protocol can be configured to run through the Pipeline Pilot Web Port so that experimental scientists only need to select file locations, inputs for the model, and target columns. A simple report dashboard is produced after each Bayesian Optimization loop so that scientists can view the progress of each iteration towards the optimum reaction conditions (Figure 4).
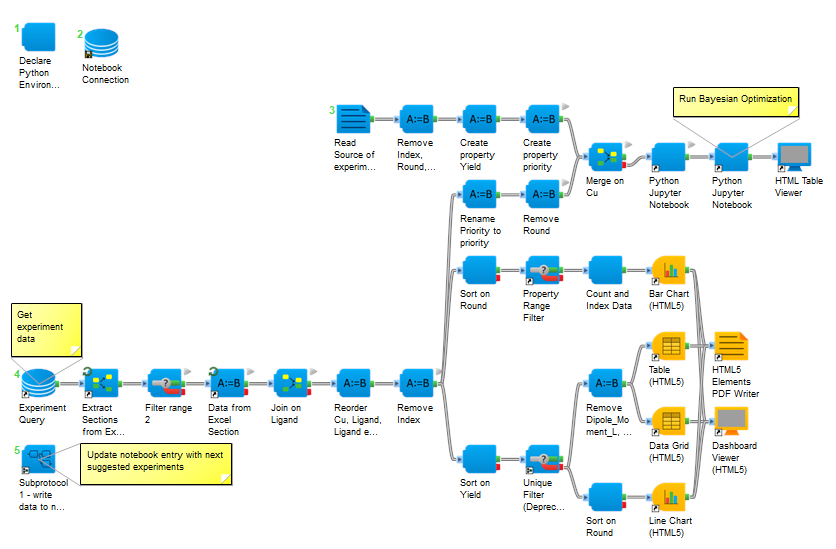
Figure 3: Bayesian Optimization with DFT molecular features.
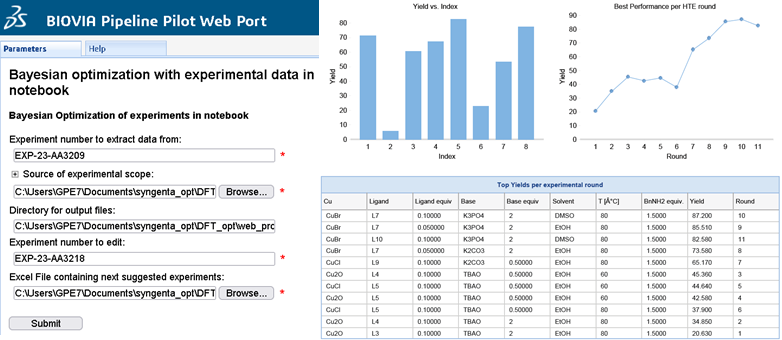
Figure 4: Example of web protocol and report dashboard.
Conclusion
A range of machine learning techniques are being applied to chemical reactions with the goal of augmenting experimental discovery and process development. Most of the open-source code is developed in Python and requires coding proficiency. BIOVIA Pipeline Pilot can be used to democratize these advanced machine learning methods providing scientists with no coding expertise the ability to deploy the latest techniques in their work.
References
(1) Ishii, A.; Kamijyo, R.; Yamanaka, A.; Yamamoto, A. BOXVIA: Bayesian Optimization Executable and Visualizable Application. SoftwareX 2022, 18, 101019.
(2) Nambiar, A. M. K.; Breen, C. P.; Hart, T.; Kulesza, T.; Jamison, T. F.; Jensen, K. F. Bayesian Optimization of Computer-Proposed Multistep Synthetic Routes on an Automated Robotic Flow Platform. ACS Cent. Sci. 2022, 8 (6), 825–836.
(3) Hickman, R. J.; Aldeghi, M.; Häse, F.; Aspuru-Guzik, A. Bayesian Optimization with Known Experimental and Design Constraints for Chemistry Applications. Digit. Discov. 2022, 1 (5), 732–744.
(4) Torres, J. A. G.; Lau, S. H.; Anchuri, P.; Stevens, J. M.; Tabora, J. E.; Li, J.; Borovika, A.; Adams, R. P.; Doyle, A. G. A Multi-Objective Active Learning Platform and Web App for Reaction Optimization. J. Am. Chem. Soc. 2022, 144 (43), 19999–20007.
(5) Shields, B. J.; Stevens, J.; Li, J.; Parasram, M.; Damani, F.; Alvarado, J. I. M.; Janey, J. M.; Adams, R. P.; Doyle, A. G. Bayesian Reaction Optimization as a Tool for Chemical Synthesis. Nature 2021, 590 (7844), 89–96.
(6) Wang, Y.; Chen, T.-Y.; Vlachos, D. G. NEXTorch: A Design and Bayesian Optimization Toolkit for Chemical Sciences and Engineering. J. Chem. Inf. Model. 2021, 61 (11), 5312–5319.
(7) Häse, F.; Roch, L. M.; Kreisbeck, C.; Aspuru-Guzik, A. Phoenics: A Bayesian Optimizer for Chemistry. ACS Cent. Sci. 2018, 4 (9), 1134–1145.
(8) Pipeline Pilot. https://www.3ds.com/products-services/biovia/products/data-science/pipeline-pilot/.
(9) BIOVIA Notebook. https://www.3ds.com/products-services/biovia/products/laboratory-informatics/electronic-lab-notebooks/biovia-notebook/.
(10) Braconi, E.; Godineau, E. Bayesian Optimization as a Sustainable Strategy for Early-Stage Process Development? A Case Study of Cu-Catalyzed C-N Coupling of Sterically Hindered Pyrazines. ACS Sustain. Chem. Eng. 2023, 11, 10545–10554.
(11) BIOVIA Pipeline Pilot Solvation Chemistry Collection.
(12) Materials Studio Collection. https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-materials-studio/materials-studio-collection/.
‡ Doyle and co-workers have released a user-friendly web version of EDBO+ which is free for academic users.

