Introduction
Glioblastoma (GBM) is the most common and deadly primary malignant brain cancer, with a 5-year mortality of over 95%, highlighting the need to identify new therapeutic solutions. Compared to the long time for developing new molecules, the repurposing of approved therapeutics has the advantage to be a quick approach. GBM is a very heterogeneous cancer with many different cell types demonstrating remarkable genomic and phenotypic variation within tumor cell populations. Therefore, modeling this heterogeneity has important implications for understanding the disease and evaluating the effect of potential therapies.
We have implemented an organoid model [1, 2] spanning multiple scales:
- an intracellular scale described by a Boolean network;
- a population scale described by an Agent-Based Model (ABM).
We use the BIOVIA Living Map app to assemble a Boolean intracellular signaling pathway encompassing the genomic and phenotypic behavior of characteristic glioblastoma (GBM) cell types.
We have computed individual cell phenotypes (e.g. death, proliferation…), fed an Agent-Based Model to describe the tumor’s progression and heterogeneity at the tissue level (figure 1). The ABM model built using our proprietary PopCell framework, highlights the variations among cell type dynamics in GBM and may be used to carry out in silico drug screenings.
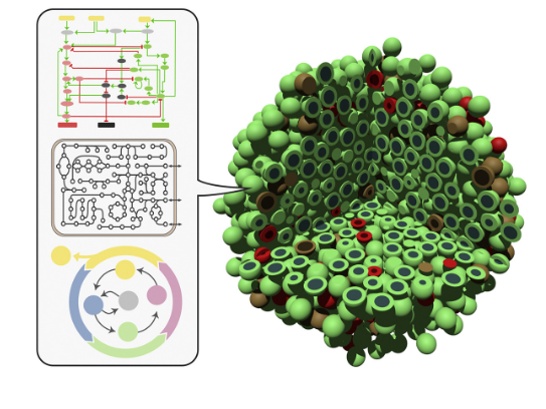
Glioblastoma Medical Need and Motivation for Digital Organoid Twins
The most common and deadly primary malignant brain cancer, glioblastoma is characterized by strong inter and intra tumor heterogeneity (figure 2), which makes finding an efficient treatment especially difficult [3].
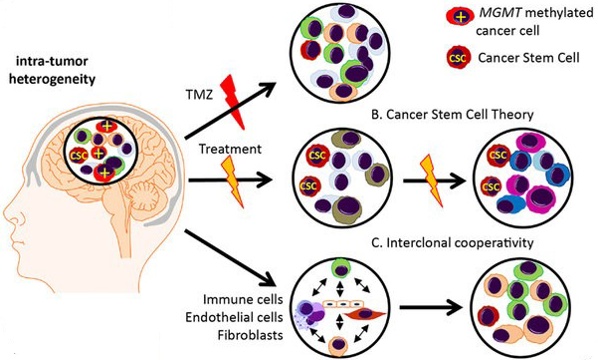
Homogeneous experimental in vitro/in vivo systems do not account for this diversity and complexity [4, 5]. Organoids may offer good experimental testbeds to tackle such issues, but they are complex to grow and depend on fresh material availability [6, 7]. A digital twin of organoids [1] may accelerate the discovery of new treatments.
Boolean Modeling
Living Map implements mathematical representations of signaling pathways where biological entity states can either be active or inactive based on their initial state value and the influence of incoming interactions. For more details on Living Map and Boolean modeling, visit our blog on Alzheimer’s Disease.
GBM Pathway
We have built 8 individual pathways, either reconstructed from published articles or imported from Pathway Databases, then merged them into a single model containing 218 entities and 417 interactions (figure 3).

We have then parametrized the pathway to mimic a variety of cell types harboring the most common mutational features of GBM cells (table1). For instance, in cell line 42-MG-BA, gene p14ARF is knocked-out and remains inactive in all simulations, while gene PDGFR is knocked-in and remains permanently active.

Drug Repurposing to Treat GBM
Developing a new drug from scratch is a very lengthy and costly process, with a high attrition rate; for any 10 drugs under development, only one will make it to market eventually. A trending strategy is therefore to repurpose existing drugs, i.e. use drugs that have already been approved for certain indications to treat another pathology. Read more on the blog: Repurposing Safe Drugs with BIOVIA Living Map.
Pathway Enrichment and In Silico Screening
Once the GBM model described above has been calibrated using validation facts from the literature, we have enriched it with 46 drugs gathered from existing databases such as IUPHAR and from scientific publications [8, 9]. We then screen drug combinations (from 1 to 3 drugs, leading to 19649 different combinations), looking for the ones that satisfy at least 6 out of 9 expected clauses (desired states of various phenotypes at the end of simulation, figure 4).

On average, we find approximately 95 different tritherapies for each cell line that satisfy 6 out of 9 clauses.
Blood Brain Barrier Penetration and Adverse Effect Prevention
One of the main obstacles to efficient chemotherapy against glioblastoma is the Brain Blood Barrier (BBB): blood vessels act as very potent filters limiting the penetration of large molecules into brain tissues. We have used a Pipeline Pilot protocol to check the BBB penetration of the various drugs in the candidate tritherapies (figure 5). Furthermore, since several drugs are administered at once, we must also check for possible adverse effects arising from drug interactions. This is done automatically with Pipeline Pilot by querying a drug-drug interaction Thesaurus (figure 6).


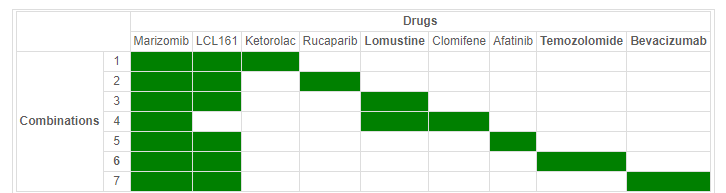
Out of the 95 tritherapy candidates, we eventually retain 7 drug combinations for cell line 42-MG-BA for instance (Table 2), some of them including lomustine, bevacizumab and temozolomide that are current chemotherapy therapeutic procedures.
We have screened drugs on all eight cell lines defined previously and identified a tritherapy (temozolomide, marozomib and LCL161) that appears to work on all of them. Marizomib (already used against multiple mieloma) is currently undergoing a phase III clinical trial as a possible treatment against glioblastoma, the result of which is due in July 2023.
Agent-Based Modeling
Using Living Map, we have built a Boolean model to study eight different cell lines appearing in glioblastoma and have successfully identifed a drug combination that seems to efficiently fight them all. However, our Boolean simulations only consider each cell line on its own. Having different cell types in a single tumor may lead to some unexpected cell interactions, especially in the presence of external stimuli such as drug combinations. In order to try and understand cell interactions, we have decided to model a population of cells using PopCell. PopCell simulates the actions and interactions of autonomous individual cells in order to understand the behavior of a system as a whole.
As a first step, we have built a simple population model where cells can divide, mutate, migrate and die (figure 7). We do not consider cell-cell interactions yet or cell-environment interaction (these will be added in future work). The various cellular processes (Division, Mutation, Migration and Death) are stochastic events governed by the outcome of the intracellular model and cell type specific parameters.

Multiscale Simulation Results
In a first set of experiments, we have simulated the growth of a glioblastoma tumor starting from stem cells. For simplicity’s sake, we have limited our simulations to a 2-dimensional grid and considered only 4 mutant cell lines (figure 8), each with its own behavior parameter to control the various cellular processes.

After a few days, the initial stem cells spontaneously mutate into new cell lines that proliferate at different speeds. Each lineage having its own characteristics, it would be interesting to add the diffusion of possible therapies (identified using our Boolean Network model,) into the model.
Conclusion and Perspectives
We successfully integrated Living Map and PopCell capabilities to create a multiscale model of GBM growth. The intracellular dynamic model with Boolean pathways in Living Map has enabled us to screen drugs and identify a promising tritherapy that acts against all the main cell lines observed in glioblastoma. The cell population model In PopCell has demonstrated the emerging heterogeneity in a glioblastoma tumor. This multiscale approach could be applied to understand other pathologies.
The current model still lacks proper modeling of cell-cell and cell-environment interactions. Further parameter refinement and model validation remain to be carried out with experimental results. This project will hopefully be continued as a PhD project (currently under construction), stay tuned!
Acknowledgements
Special thanks to our former intern Hala Bouzidi who built the models and did the bulk of the work reported here.

