
Recycling plastic is a crucial aspect of sustainability, but remains a challenging task.
Mechanical recycling, where the plastic is broken down mechanically, is one of the most common forms of recycling but, with repeated application, leads to products with degraded properties. Currently, chemical recycling is popular but is not cost-effective compared with creating virgin polymers – so can we use enzymes to break down or compost plastics?
Compostable plastics such as Polylactic acid (PLA) require an industrial composter to break down due to the high temperatures involved. Even then, polymers like PLA take several weeks to break down and this is not financially viable. Some polysaccharide based materials can degrade significantly faster, but there are challenges with processing and manufacturing these materials.
Experiments have been carried out on similar polymers where they embed the digester enzyme in the polymer and activate it to break down the polymer from the inside rather than outside. This should lead to faster degradation than with existing methods. The question is how to embed the enzyme so that breakdown can be controlled.
Researchers have developed random heteropolymers that mimic an enzyme’s mixture of hydrophobic and hydrophilic surface segments. These can bind to both the polymer matrix and the enzyme and hence enable the embedding of the enzyme in the polymer matrix. This enzyme can be activated by the right heat and light combination to enable on-demand breakdown of the polymer.
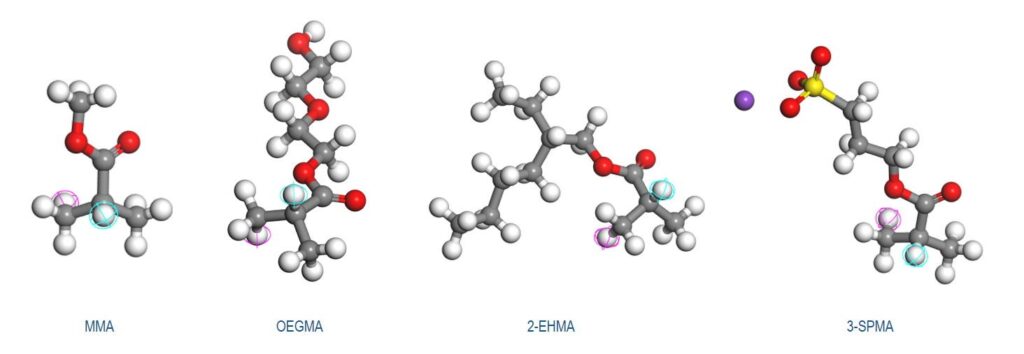
In this example, the random heteropolymers are block copolymers consisting of 4 different monomers (see Figure 1). During batch polymerization, composition drift can occur which leads to composition gradients in the copolymers. Researchers have developed a program called Compositional Drift that generates strings encoding the correct ordering of repeat units to build random heteropolymers. A MaterialsScript was written to read in the output from the program and create atomistic models of the random heteropolymers which include the composition drift.
For the simulations, a model enzyme, Burkholderia cepacia lipase (BC-Lipase) which accelerates the digestion of polyesters, was obtained from the Protein Data Bank and cleaned using the Prepare Proteins protocol in BIOVIA Discovery Studio. The enzyme was placed into a cubic cell and the BIOVIA Materials Studio Amorphous Cell solver was used to pack polymers around the enzyme. For this project, the random heteropolymer, PLA, and then other pure polymers consisting of repeat units from the random heteropolymer were included to compare with the random heteropolymer. The systems were equilibrated using several cycles of molecular dynamics with the Forcite solver running on a GPU and the COMPASS III forcefield. After equilibration and production runs, the interaction energy between the polymer and enzymes was calculated.
Simultaneously, molecular dynamics simulations of the polymers without the enzyme were executed to establish the density and solubility parameters for comparison with PLA. Note that this was a proof of concept set of calculations and simulations were not replicated multiple times. The results are shown in Figure 3.
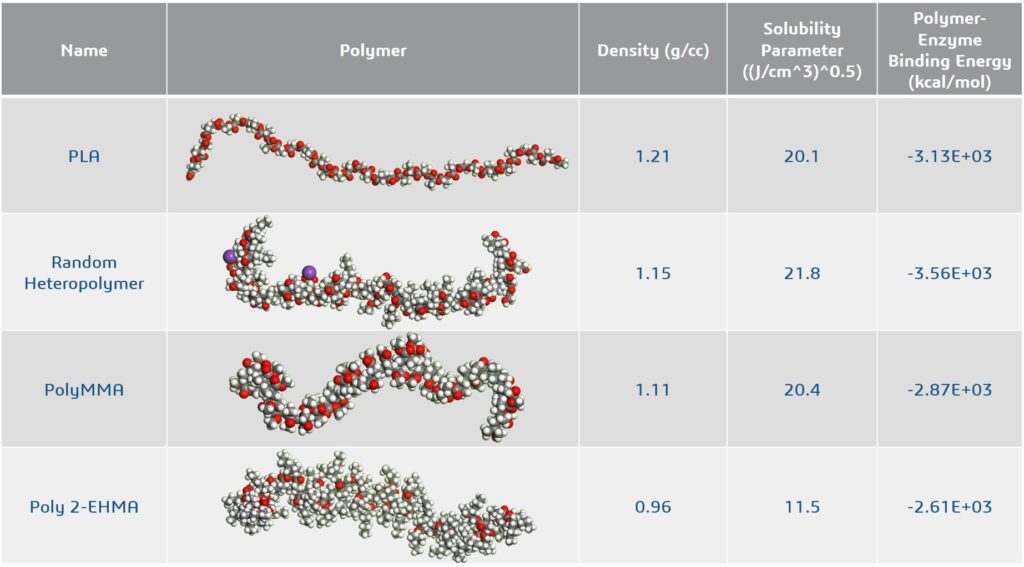
From these simple simulations of the three candidate polymers, the random heteropolymer is closest in density and solubility parameters to PLA indicating it will be most compatible with PLA. The binding energy between the enzyme and the random heteropolymer is also stronger than that between PLA and the enzyme. Hence, the random heteropolymer is likely to bind preferentially to the enzyme.
Models of enzyme encapsulation by random heteropolymers have been developed using virtual experiments.
Further virtual experiments could be used to optimize the repeat unit ratios in the random heteropolymer to design improved systems for controlled degradation of polymers.
Want to learn more about the end-to-end strategy that delivers sustainable packaging with speed, quality and cost efficiency?

