AI for the Biologist
Only within the last couple of years, we have witnessed how artificial intelligence (AI) is reshaping the way biological research is done. While experimental validation remains essential, AI now allows us to predict protein structures directly from amino acid sequences — a breakthrough made possible by the wealth of high-quality data in the Protein Data Bank (PDB). Beyond structure prediction, AI can even generate entirely new proteins from scratch, as demonstrated by recent work from the Baker Lab and others. More recently, the emergence of co-folding models such as AlphaFold3 and OpenFold3 represents a significant leap forward, enabling the direct prediction of the structures of multimolecular complexes involving proteins, nucleic acids, and even small molecules from sequence information. All these advances dramatically expanded the landscape of rational design, opening new avenues for molecular design and drug discovery.
While the opportunities that these AI-driven tools present are incredibly exciting, computational biologists often find themselves searching for the best and the most efficient strategy to integrate them into their protein engineering workflows. RFdiffusion, ProteinMPNN, and AlphaFold2 are highly useful for de novo protein design; yet, how a modeler might approach design campaigns with such tools can be challenging. For instance, the design of a protein binder to a specific protein target typically involves multiple stages: (1) generating potential scaffolds for the binder, (2) assigning sequences to these scaffolds, and (3) building the structures based on these sequences. Throughout the design process, multiple parameters control each stage. Moreover, an optimal set of parameters from one design campaign may not carry over to the next. Thus, there is a demand for an integrative design process that can explore a large parameter space.
Solving the Challenge: One Protocol to Rule Them All
With the latest version of Discovery Studio Simulation on 3DEXPERIENCE® platform, we introduce to our users a new protocol, Design Protein Binders, that addresses these needs for de novo binder design. Design Protein Binders leverages RFdiffusion to generate scaffolds for a binder to the target protein, ProteinMPNN to generate optimized amino acid sequences for these scaffolds, and AlphaFold2 to predict the structure of each sequence and evaluate the structural similarity to the scaffold from RFdiffusion.
In the first stage of the de novo binder design pipeline, Design Protein Binders uses a sequential design process to explore a user-defined parameters space and design a new protein predicted to bind a known target protein structure. In the second stage of the pipeline, Design Protein Binders uses the same sequential design process to sample similar interfaces to the top designs from the first stage. Specifically, residues distal to the binding interface are partially diffused to sample additional protein scaffolds that preserve the binding interface. In both the first and second stages of the design pipeline, users can set filter criteria for design scores such as predicted local distance difference test, predicted aligned error, interface predicted aligned error, and root-mean-square deviation between the AlphaFold2 predicted structure of the complex relative to the scaffold designed by RFdiffusion. Designs that don’t meet filter criteria are automatically pruned. The result is an automated de novo design process that users can further modify to adjust the exhaustiveness of the design process, (see figure).
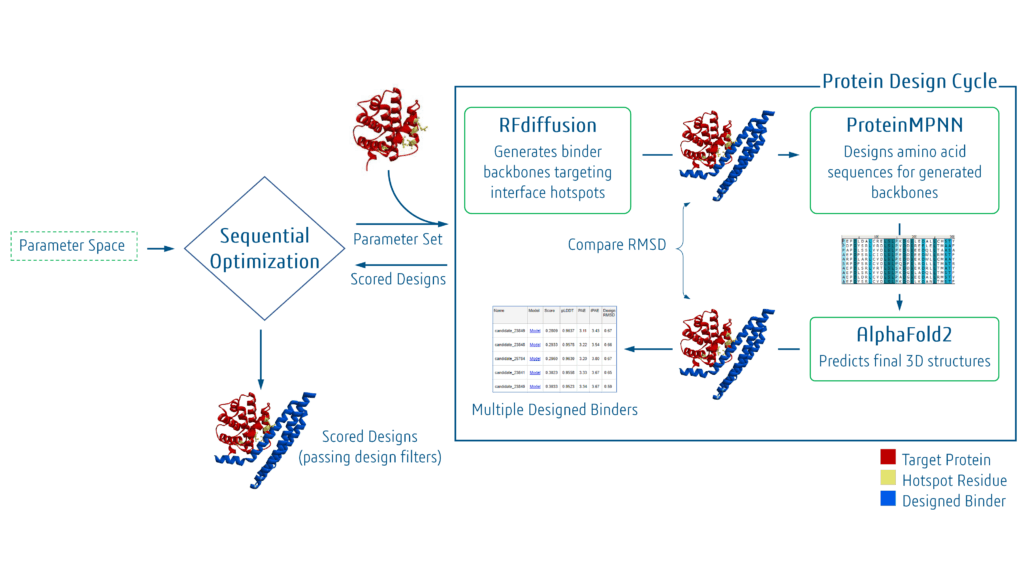
The Design Protein Binders protocol efficiently samples thousands of binder designs across a user-defined parameter space by omitting the multiple sequence alignment generation step from the structure prediction. Users can also calculate score terms automatically with additional steps in BIOVIA Discovery Studio Simulation. Users also have the option to further refine top binder candidates manually or confirm the predicted structure with multiple sequence alignments with the existing Generate Protein Scaffolds, Generate Protein Sequences, and Predict Protein Structures protocols.
The Design Protein Binders protocol represents a significant leap forward for de novo protein design in Discovery Studio Simulation, providing researchers with an efficient, intelligent, and flexible platform to accelerate the discovery of ground-breaking new protein binders. This protocol complements the separate pipelines for generating protein scaffolds with RFdiffusion, for generating sequences for a protein scaffold, and for predicting the structure of a protein sequence already in BIOVIA Discovery Studio Simulation, and may yet prove to be the protocol to bring all these tools together.
-Tien Luu, Ph.D.Portfolio Senior Manager, Biosciences – Dassault Systèmes, BIOVIA
As scientific AI continues to transform rational design of proteins, integrated workflows help accelerate discovery by democratizing these sophisticated strategies to biologists. BIOVIA is leading the way, making AI-driven protein design accessible, efficient, and actionable.
Watch the video to learn more about our new Design Protein Binders protocol:
📩Want to find out the latest news about BIOVIA events, customer stories, blogs and more? Join the newsletter today!